Abstract. The circumscription and relationships among members of Comaceae have long been controversial. Seventeen diverse genera (Cornus, Alangium, Nyssa, Davidia, Camptotheca, Diplopanax, Garrya, Mastixia, Curtisia, Corokia, Helwingia, Aucuha, Griselinia, Melanophylla, Aralidium, Toricellia, and Ka-liphora) have been ascribed to this family by various authors. Parsimony analyses of rbcL sequences of all 17 genera, as well as other putative relatives, revealed a cornaceous clade consisting of Cornus, Alangium, Nyssoids (Nyssa, Davidia, Camptoiheca), Mastixioids (Diplopanax, Mastixia), Curtisia, Hydrangeaceae, and Loasaceae. This rbcL based cornaceous clade differs from all previously proposed comaceous groups. Within this cornaceous clade, four major lineages were identified: i) Cornus-Alangium, ii) Nyssoids-mastixioids, iii) Curtisia, and iv) Hydrangeaceae-Loasaceae. Significantly, rbcL sequence data also revealed that some genera (Corokia, Helwingia, Aucuba, Garrya, Griselinia, Melanophylla, Kaliphora, Aralidium, and Toricellia) previously placed in Comaceae by some authors as well as some genera (Quintina, Abrophyllum, Polyosma, and Escallonia) traditionally allied with Hydrangeaceae are phyloge-netically distantly related to the rbcL-based cornaceous clade.
Key words. RbcL sequence data, phylogeny, Cornaceae s. l.
Comaceae sensu lato were described as a "dustbin" by Eyde (1988) because of the morphologically diverse array of taxa ascribed to the family. Fifteen genera were attributed to Cornaceae following the broadest definition (Harms, 1898); a total of 17 genera has been placed in the family when the treatments of all authors are considered. Despite its small size, however, the morphological heterogeneity among members of Cornaceae (Harms, 1898) is so great that most of the genera originally placed in Comaceae by Harms have been treated either as separate families or orders, or as members of other families (Table 1). The 17 genera considered members of Cornaceae by different authors at one time or another are: Alangium, Aralidium, Aucuba, Camptotheca, Cornus, Corokia, Curtisia, Davidia, Diplopanax, Garrya. Griselinia, Helwingia, Kaliphora, Mastixia, Melanophvlla, Nyssa, and Toricellia. These 17 genera will hereafter be referred to as Comaceae s. l. Most of these genera are small (one to five species) and geographically isolated (reviewed in Xiang et al., 1993). Only the genera Alangium, Aucuba, Cornus, Garrya, and Mastixia have more than ten species. Ten of these 17 genera are found in the Sino-Japanese flora (Alangium, Aucuba, Camptotheca, Cornus, Davidia, Diplopanax, Helwingia, Mastixia, Nyssa, and Toricellia).

Although most of the 17 genera of Comaceae s. l. share certain features (e.g., small 3-5 merous flowers having an inferior ovary, epigynous disk, one ovule per locule, and a drupaceous fruit) they also exhibit a wide range of variation in morphology. For example, Helwingia has stipules and unisexual flowers that are epiphyllous whereas the other genera do not possess stipules and have bisexual or unisexual flowers arranged in axillary or terminal inflorescence of various types. Similarly, Nyssa, Aralidium, Melanophylla, and Griselinia have imbricate petal aestivation but the remaining genera all have valvate petals or no petals.
Corokia is unique in Cornaceae because of its long and short shoots with the leaves clustered on the latter. Garrya is also distinctive in that it possesses slender axillary catkins arranged in whorls subtended by opposite bracts.
This great morphological diversity has resulted in tremendous difficulty in defining Cornaceae. As a result, many classifications of Cornaceae have been proposed. A partial list (Table 1) includes Harms (1898), Wangerin (1910), Hutchinson (1967), Takhtajan (1987). Cronquist (1988), Eyde (1988), and Thorne (1992). These schemes differ from one another not only in how they define Cornaceae, but also in how they ally the family with other families. The magnitude of controversy regarding the circumscription of the family can be well illustrated by the treatments of Cronquist (1988) and Takhtajan (1987) (Table 1). Cronquist (1981) largely followed Wangerin (1910) and included ten genera (Mastixia, Curtisia, Toricellia, Helwin-gia, Aucuba, Kaliphora, Carnus, Corokia, Griselinia, and Melanophylla) in his Cornaceae and later (1988) added the nyssoids (i.e. Nyssa, Davidia, and Camptotheca) to Cornaceae following Eyde (1988), as well as Aralidium. In the most narrow treatment yet proposed for Cornaceae, Takhtajan (1987) treated many of the genera originally placed in Cornaceae as distinct families and retained only Cornus in the Linnaean sense (1753). For a more detailed discussion of the taxonomic schemes proposed for Cornaceae, see Xiang et al. (1993).
To understand better the circumscription and relationships of Cornaceae, we employed comparative gene sequencing, which has shown great potential for solving problems at a wide range of taxonomic levels depending on the gene or gene segment employed (reviewed in Clegg & Zurawski, 1992; Palmer, 1987; Palmer et al., 1988). One gene in particular, the chloroplast gene rbcL (coding for the large subunit of ribulose-1.5-bisphosphaie carboxylase/oxygenase), has been particularly useful in elucidating phylogenelic relationships of plants at the generic level and above (e.g. Chase et al., 1993; Giannasi et al., 1992; Kron & Chase, 1993; Morgan & Soltis, 1993; Olmstead et al.. 1992, 1993; Qiu et al., 1993, Rodman et al., 1993; Soltis et al., 1990, 1993; Xiang et al., 1993). Advantages of rbcL sequence data for retrieving phytogeny include a rate of sequence evolution conservative enough for comparisons at higher taxonomic levels, the presence of rbcL as a single-copy gene (see Chase et al., 1993 for examples of rbcL duplication), and the abundance of ehloroplast DNA (reviewed in Clegg & Zurawski, 1992; Palmer et al., 1988). In a recent study using comparative sequencing of rbrL, Xiang et al. (1993) examined 13 of the 17 putative corna-ceous genera and showed that rbcL sequence data are extremely useful in unraveling the taxonomic problems surrounding Cornaceae. Xiang et al. (1993) not only demonstrated that most previously defined Cornaceae are either polyphyletic or paraphyletic, hut also showed that genera from Hydrangeaceae (Hydrangea, Philadelphus, Deutzia, Decumaria, and Carpenteria), as well as Atangium, Camptotheca, Cornus, Curtisia, Davidia, Diplopanax, Mastixia, and Nyssa constitute a cornaceous clade. Furthermore, Xiang et al. showed that several genera previously placed in Cornaceae by some authors are only distantly related to this comaceous clade (e. g., Aucuba, Corokia, Garrya, Griselinia, Helwingia).
Because Xiang et al. (1993) demonstrated a close relationship between Cornaceae and Hydrangeaceae, it is necessary to analyze proposed relatives of Hydrangeaceae to determine if these taxa are also part of the comaceous clade. Recently, Hufford (1992) and Downie & Palmer (1992) proposed a close relationship between Loasaceae and some members of the cornaceous clade (e.g., Hydrangeaceae and Cornaceae) based on morphological and chemical data, and restriction site mapping of the chloroplast genome inverted repeat, respectively. Takhtajan (1987) also considered several other families closely related to Hydrangeaceae and Cornaceae (e.g., Escalloniaceae, Abrophyllaceae, Argophyllaceae, Polyosmataceae). Morgan and Soltis (1993) sequenced rbcL from a species of Escallonia. but other Escalloniaceae, as well as Abrophyllaceae, Argophyllaceae, and Polyosmataceae need to be investigated in this regard. In this paper, we conducted a broader phylogenetic analysis of rbcL sequence data than that conducted by Xiang et al. (1993) that included all putative genera of Cornaceae s. l., as well as potential relatives of Hydrangeaceae and Cornaceae not previously analyzed (members of Loasaceae, Abrophyllaceae, Polyosmataceae, and Escalloniaceae) with two major objectives: 1) better define the comaceous clade and 2) elucidate the relationships within the comaceous clade.
Materials and Methods
We generated rbcL sequences for Aralidium, Kaliphora, Melanophylla, and Toricellia (Table 2), four proposed members of Cornaceae not sequenced earlier by Xiang el al. (1993). These sequences, plus those of Xiang et al. provide data for all 17 genera included in Cornaceae by at least one author. RbcL sequences of five proposed relatives of Hydrangeaceae [Petalonyx and Mentselia (Loasaceae), as well as Polyosma (Polyosmataceae), Abrophyllum (Abrophyllaceae), and Quintinia (Escalloniaceae)] were also included in this study. We generated rbcL sequences for all of these genera (Table 2), except Mentzelia, the sequence of which was kindly provided by R. G. Olmstead (unpublished).
Plant material for sequencing was mostly obtained from fresh leaves or silica-dried leaves, or in the case of Melanophylla, from leaves of a recently collected herbarium specimen (Table 2). Methods used for DNA extraction, gene amplification, sequencing, and information regarding primers were described earlier (e. g., Morgan & Soltis, 1993; Xiang et al., 1993). Sequences were typically amplified using PCR primers Z 1 (forward primer) and 3' rbcL (reverse primer). For two taxa, Toricellia and Melanophylla, we were unable to amplify rbcL using this Z 1-3' rbcL primer combination. For these genera, we therefore used an internal reverse primer (Z 1351 R) in place of the 3' rbcL primer, a method we employed earlier for Masima and Diplopanax in Xiang et al. (1993).
Our broad parsimony analysis included 50 sequences of rbcL. In addition to the sequences noted above, rbcL sequences of other representatives of Asteridae s. l. were included. These sequences are from Chase et al. (1993), Morgan and Soltis (1993), and Olmstead et al. (1993) (see Appendix to Annals of Missouri Botanical Garden Vol. 80, No. 3). To be consistent, those taxa used as outgroups (Cercidiphyllum, Itea, Saxi-fraga, Asfibe, and Daphniphyllum) in the analysis of Xiang et al. (1993) were also chosen as outgroups for this broad analysis. A total of 1377 bp of rbcL (bases 31-1407) was standardly used for this analysis. For Mastixia, Diplopanax, Toricellia, and Melamphylla, 1320 bp of rbcL (bases 31-1350) were used due to the different strategy employed for the rbcL amplification for these four genera (see above and Xiang et al.,1993).
Sequences for the broad analysis were analyzed using PA UP version 3.1.1 (Swofford, 1993). A single search with MULPARS using simple taxon addition and tree-bisection-reconnection (TBR) branch-swapping with characters specified as unordered and unweighted was first conducted. One hundred consecutive searches were subsequently performed with MULPARS using random taxon addition and TBR branch-swapping to determine if several islands of minimal-length trees exist (Maddison, 1991). This broad parsimony analysis revealed a cornaceous clade of 15 genera (see results). To evaluate the relative robustness of the cornaceous clade and other clades found in the most parsimonious trees, a bootstrap analysis (Felsenstein. 1985) with 100 replicates was initiated with simple taxon addition, TBR branch-swapping, and MULPARS, but was stopped at replicate 62 due to the tremendous amount of time this required (48 days). Due to the memory limit of PAUP (only 32,767 trees can be saved, Swofford, personal communication), a decay value (Donoghue et al., 1992) could be obtained for the comaceous clade only by following the general method of Johnson and Soltis (1994). This approach involves using a strict consensus tree of all most parsimonious trees as a constraint tree for heuristic searches that save only trees of one additional step that fail to satisfy the constraint topology.
Detailed phylogenetic analyses were subsequently performed only for members of the comaceous clade recognized by the broad analysis described above. This second analysis included 24 taxa (15 taxa included in the initial broad parsimony analysis, as well as seven and two additional taxa from Comus and Hy-drangeaceae, respectively) and was conducted to elucidate relationships within the comaceous clade. A total of 1440 bp (bases 31-1470) was used for the detailed analyses of the comaceous clade. Diospyros, Sar-racenia, Roridula, and Fouquieria were chosen as outgroups for these detailed analyses based on the results of the big analysis of seed plants by Chase et al. (1993). In Chase et al. these four genera were phylo-genetically close to the cornaceous clade. Sequence data were analyzed using PAUP 3.1.1 as described above, except that 100 replicates were completed in the bootstrap analysis; furthermore, decay values were obtained for all clades recognized in the most parsimonious trees by examining trees up to five steps longer than the shortest.
Results
1. RbcL sequence divergence
Among the 17 sequences of Cornaceae s. l., 282 sites of rbcL are variable (19.7%) and 126 sites are phylogenetically informative (8.8%). Among the 24 sequences of the cornaceous clade, 237 sites are variable (16.5%) and 117 sites are informative (8.1%). These data show that sequence divergence is higher among the 17 putative genera of Cornaceae s. I. than it is among the comaceous clade. The sequences obtained for the region flanking the 3' end of rbcL are more variable than the coding region, which agrees with the finding of Soltis el al. (1993) for Saxifragaceae s. s. and Morgan et al. (1994) for Rosaceae. This flanking region appears to evolve about one and half times as fast as the coding region. In the comaceous clade, nine out of 42 bp are variable (21.4%) in the region flanking the 3' end of rbcL and only one of these variable sites is phylogenetically informative (2.4%).
2. RbcL. structural changes
There are no deletions or insertions found before base 1421 among the rbcL sequences of Comaceae s. l., Hydrangeaceae, and putative relatives of Hydrangeaceae included in this study. Different structural changes at the 3' end of the gene are found in several taxa that are not members of the cornaceous clade: Griselinia, Abrophyllum, Corokia, Aralidium, Polyosma, and Kaliphora. A fragment of 11 bp (GGATTAGTTGT) is inserted between bases 1224 and 1425 in Grisellnia. This insertion renders the original termination codon (bases 1426-1428) nonfunctional, hut creates a new stop codon at position 1429, increasing the effective length of rbcL in Griselinia by one codon (Fig. 1 a). In Abrophyllum, a fragment of 16 bp (1426-1441) apparently is deleted beginning at the termination codon and replaced by a fragment of 40 bp (GATCGCCAAGAATCCGGTGCATTGCCATCCGATAAATAAA), creating a new stop codon at position 1462 (Fig. 1 b), thus increasing the effective length of rbcL by 36 bp. In Polyosma, an insertion of 19 bp occurs between bases 1421 and 1422, four base pairs before the original stop codon. However, the insertion creates a new stop codon at the same position as the original (bases 1426-1428), thus the effective length of rbcL remains 1428 bp (Fig. 1 e). In Kaliphora, an insertion of 39 bp (AGATCCGAAGACCAAAGATCCGAAAGATCCGAAGACCCA) occurs between bases 1424-1425, increasing the effective length of rbcL by 13 codons (Fig. 1 h). Bases 1429-1436, and 1454-1458 are deleted in Kaliphora (Fig. 1 h).
In Corokia and Aralidium structural changes occur downstream of the termination codon, and thus do not affect the length of rbcL. In Corokia, bases 1429-1460 are deleted and replaced by a large fragment of 99 bp (Fig. 1 c). The first 42 bp of this fragment represent a duplication of bases 1380-1421, with G to A base changes at positions 1435 and 1465 (Fig. l c). In Aralidium, a deletion of five base pairs (1459-1463) occurs in the region flanking the 3' end of the rbcL (Fig. 1 d).

Two structural changes are also observed in members of the cornaceous clade, one involving Loasaceae and a second one present in two Cornus species. The effective length of rbcL in all members of the comaceous clade is 1428 bp, except in Petalonyx and Mentzelia (Loasaceae), in which rbcL is six base pairs longer due to an insertion (GGATCA) between bases 1424 and 1425, making rbcL 1434 bp (Fig. 1 f). This structural change occurs only in Petalonyx and Mentzelia and provides additional support for a close relationship between these two genera of Loasaceae. An additional three base pairs (TAA) are also inserted four base pairs downstream of the termination codon in Petalonyx and 8 bp are deleted after the termination codon in Mentzella.
Another structural change is found in the two Asian blue-fruited species of Cornus sequenced (C. ob-longa and C. walferi). In both of these species, bases 1379-1421 (25 bp: TATTTGAATTCGAAGCAATGGATAC) are repeated. Furthermore, a base substitution (from A to T) has apparently occurred at base position 1422 in both species (Fig. 1 g). This duplication, however, does not affect the length of rbcL because it creates a new stop codon at the same position (bases 1426-1428) as in species that do not have the duplication (Fig. 1 g). This structural change is not present, however, in the North American bine-fruited species of Cornus sequenced (C. obliqua and C. alternifolia).
3. Parsimony analyses
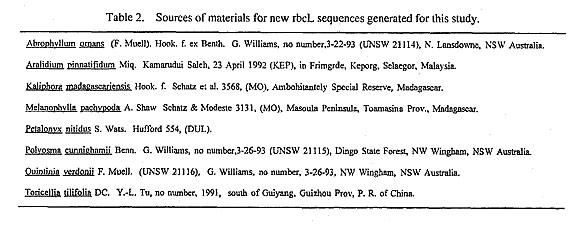
a. The broad analyses. The single search involving the large data matrix with simple taxon addition resulted in 214 most parsimonious trees each of 1419 steps and with a consistency index of 0.436. The subsequent 100 consecutive searches with random taxon addition found 449 trees of 1419 steps in four islands. The first island contained 214 trees and was the one found by the single search with simple taxon addition, This island was found 68 times. The second island contained six trees and was found seven times. The third island contained 219 trees and was found 20 times. The fourth island contained 10 trees and was found five times.
Differences among the topologies of the strict consensus trees of each of the four islands mainly involve the placements of two genera of Escalloniaceae (Escallonia and Quintinia), Polyosma, Berzelia, and Viburnum (Fig. 2). All of the 449 equally most parsimonious trees recognized three major clades (Fig. 2): 1) a cornaceous clade consisting of four major lineages [nyssoids-mastixioids (Mastixia and Diplopanax); Comus-Alangium; Curtisia; and Hydrangeaceae-Loasaceae]. 2) a clade consisting of Diospyros, Rhododendron, and Styrax, and 3) a clade of traditional asterids, Apiales, and other putative relatives of Cornaceae and Hydrangeaceae.
Some genera considered members of Cornaceae or possible close relatives of Cornaceae or Hydrangeaceae, such as Kaliphora, Aucuba, Garrya, Quintinia, Abrophyllum, Corokia, Griselinia, Aralidium, Toricellia, Melanophylla, Helwingia, and Polyosma, are not members of the cornaceous clade, but are instead allied with a wide array of taxa in the Asteridae s. l. (Fig. 2). For example, in this analysis Kaliphora appears closely related to a higher asterid, Nicotiana; Aucuba and Garrya are closest relatives and appear as the sister to Eucommia; Corokia and Argophyllum are in a clade with two higher asterids, Helianthus and Lobelia; Griselinia, Aralidium, Toricellia. and Menalophylla are closely allied with representatives of Apiales; Helwingia is allied with Phyllonoma; Quintinia, Escallonia, and Polyosma appear at or near the base of a large clade containing Griselinia, Toricellia, Melanophylla, Aralidium, and Apiales.
The cornaceous clade was present in all trees up to two steps longer than the most parsimonious trees (with a decay index of 3). The bootstrap analysis provided strong support for the cornaceous clade (70% bootstrap value) (Fig. 2). Other strongly supported clades recognized in the broad analysis are the sister group relationship of Aucuba-Garrya clade (95% bootstrap value and 10 nucleotide substitutions), and the Helwingia-Phyllonoma-Ilex clade (92% bootstrap value and 10 nucleotide substitutions).
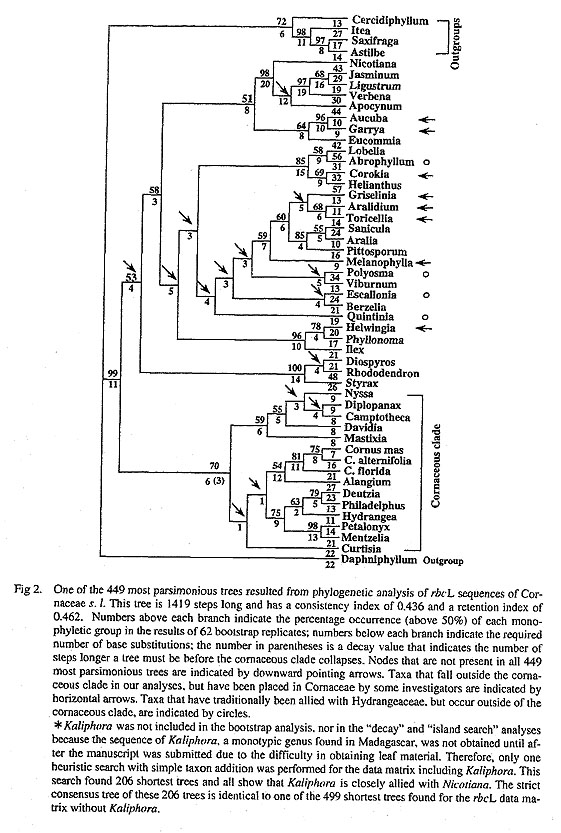
b. Analyses for the cornaceous clade. The single search with simple taxon addition for the comaceous clade using a total of 1440 bp (which includes the coding region of rbcL and the region flanking the 3' end of the gene) yielded 97 most parsimonious trees each of 568 steps and with a consistency of 0.62. The subsequent 100 consecutive searches with random taxon addition found the same 97 trees of 568 steps, suggesting that multiple islands of shortest trees are not present. All of the 97 most parsimonious trees (Fig. 3) recognized the same four major lineages within the cornaceous clade as those found in the broad analysis. The shortest trees differ in the placements involving the four major lineages. For example, some shortest trees show that Curtisia is the sister of Cornus-Alangium, nyssoids-mastixioids are the sister group of Curtisia-Cornus-Alangium, and Hydrangeaceae-Loasaceae are the sister to the remainder of the cornaceous clade (Fig. 3). However, some other equally most parsimonious trees show that Hydrangeaceae-Loasaceae are the sister group of Cornus-Alangium, with Curtisia the sister of the Cornus-Atangium-Hydraageaceae-Loasaceae clade and nyssoids-mastixioids the sister group of the remainder of the cornaceous clade. Bootstrap analyses provided poor support for the relationships among these four lineages within the cornaceous clade (50%). The strict consensus tree shows a tetrachotomy of the four groups, which agrees with Xiang et al. (1993) although Loasaceae were not included in that earlier study. The placements of taxa within clades of Comus, nyssoids, and Hydrangeaceae-Loasaceae also differ among the shortest trees. For example, some shortest trees show that the big-bracted dogwoods-dwarf dogwoods clade is basal within Cornus,
whereas in others the blue-fruited dogwoods are the basal lineage within Comus. Similarly, some shortest trees show that Hydrangeaceae and Loasaceae are each monophyletic, whereas in others Hydrangeaceae are paraphyletic with Hydrangea the sister of Loasaceae (Fig. 3). The relationships of Loasaceae to Hydrangeaceae and generic relationships within Hydrangeaceae have been examined in more detail (Soltis et al., in preparation).
Discussion
Our broad rbcL sequence analysis demonstrated that Cornaceae sensu Harms (1898), Wangerin (1910), Hutchinson (1967), and Cronquist (1988) are polyphyletic and Cornaceae sensu Eyde (1990) and Thorne (1992) are paraphyletic. These findings agree with our earlier analysis of rbcL sequences (Xiang et al., 1993). Thus, sequence data suggest that the morphological features that have been used to unite these taxa, such as inconspicuous flowers, separate petals, partially united sepals, epigynous discs, inferior ovaries, mostly solitary ovules per locule, and mostly drupaceous fruits are homoplasious. Significantly, nine of the genera (Kaliphora, Aucuba, Garrya, Corokia, Griselinia, Helwingia, Aralidium, Toricellia, and Melano-phylla) that are proposed members of Cornaceae are not members of the rbcL-based cornaceous clade, but are allied instead with a wide array of asterids. The phylogenetic positions revealed by our broad analysis
for five of these genera (Aucuba,Gurrya, Corokia, Griselinia. and Helwingia) are congruent with Xiang et al. (1993) and also Olmstead et al. (1993): Aucuba is the sister of Garrya: Corokia is a close relative of Asteraceae; Griselinla is allied with Apiales, and Helwingia is the sister of Phyllonoma. The relationships of these genera are discussed in more detail in Xiang el al. (1993). Herein we discuss only the relationships of the remaining four genera, Aralidium, Toricellia, Kaliphora. and Melanophylla.
Aralidium, Toricellia, Kaliphora, and Melanophylla are revealed by this study to be only distantly related to the cornaceous clade. Toricellia consists of two to three species in the eastern Himalaya and south western China and was placed in Cornaceae by Harms (1898), Wangerin (1910), Hutchinson (1969), and Cronquist( 1981.1988). Eyde (1988) removed it from Cornaceae because of its distinctive cordate-rounded, paimately-nerved leaves having a petiole broadly sheathing at the base, thick branches, Sambucus-tike pith, and multicellular glandular hairs. However, Eyde (1967, 1988) did find Cornus-like transsepta! bundles in Toricellia. That is, because there is no vascular tissue in the central axis of ovary, the ventral bundles rise through the ovary wall and not through the axis to supply the pendulous ovule. However, Eyde maintained that this feature had evolved independently in Toricellia and his true cornoids (Cornus, Mastixia, and nys-soids). Toricellia also has highly specialized wood that is distinct from that present in Comus (Adams, 1949; Li & Chao, 1954). Dahlgren (1975) and Thorne (1983, 1992) placed Toricellia as a monotypic family under Araliales. Takhtajan (1980) placed Toricelliaceae in Cornales and considered it as an intermediate between Cornaceae and Araliaceae. Later, Takhtajan (1987) established a separate order for Toricellia, To-ricelliales, and placed them close to his Cornales. Our rbcL sequence analyses revealed that Toricellia is clearly not a member of the cornaceous clade. but instead is closely related to Aralidium (discussed below) and allied with Apiales (Fig. 2). This sister relationship between Toricellia and Aralidium was recognized in all the 449 most parsimonious trees and supported by six nucleotide changes and a 68% bootstrap value. This relationship is also supported by a more detailed analysis of rbcL sequences for Apiales (Plunkett, personal communication). Hence, the rbcL sequence data support the treatments of Thorne (1983, 1992) and Dahlgren (1975) and also agree with Eyde's (1988) hypothesis that the Cornus-like transeptal bundles evolved independently in cornoids and Toricellia.
Aralidium, consisting of a single species in Sumatra and the Malay Peninsula, was placed in Araliaceae by Hutchinson (1967), but Cronquist (1988) moved it to his Cornaceae. Thorne (1992) separated it as a monotypic family and placed it in his Cornales. Our rbcL sequence data strongly suggest that Aralidium and Toricellia are closely related and allied with Apiales. This close relationship between Aralidium and Toricellia is also supported by several morphological characters. Both Aralidium and Toricellia have trans-septal bundles, empty ovary chambers, glandular hairs, thick branches, and leaves possessing iridoids (Eyde, 1988). Again, a more detailed phylogenetic analysis of rbcL sequences for Apiales and relatives also supports this close relationship between Aralidium and Toricellia, as well as a close relationship of these genera to Apiales (Plunkett, personal communication).
Evidence from floral anatomy (Eyde, 1967), pollen morphology (Ferguson 1977; Ferguson & Hideux, 1980), wood anatomy (Adams, 1949), and phytochemistry (Bate-Smith et al., 1975) all supported the exclusion of Kaliphora and Melanophylla from Cornaceae. However, Cronquist (1981, 1988) still placed these genera in Cornaceae. These two genera, found only in Madagascar, are usually considered to be close to each other and have been traditionally placed in Cornaceae (Harms, 1989; Wangerin, 1910). Takhtajan (1980, 1987), however, placed these two genera in his Melanophyllaceae. Pollen morphology (Ferguson 1977; Ferguson & Hideux, 1980) suggested an affinity of Kaliphora. Melanophylla and Montinia, the latter genus a traditional member of Saxifragaceae s. l. (see Morgan & Soltis, 1993). Based largely on pollen morphology. Thorne (1983) placed all three of these genera in subfamily Montinioideae (Saxifragaceae), but later (1992) recognized Montiniaceae. which he placed in his Hydrangeales (Table 1). Similarly, Takhtajan (1987) placed Melanophyllaceae in Hydrangeales next to Montiniaceae. According to rbcL se-quence data, however, Kaliphora and Melanophylla arc not closely related to Cornus and Hydrangeaceae, but are more closely allied with representative of Solanales and Apiales, respectively (Fig. 2), suggesting that Kaliphora and Melanophylla are distantly related to each other. Furthermore, analysis of rbcL sequences for Saxifragaceae s. l. (Morgan & Soltis, 1993) similarly revealed that Montinia is a close relative of Convolvulaceae and Solanales (Asteridae), and distantly related to Apiales. Although more exhaustive phylogenetic analyses of Asteridae s. I. are needed to determine the closest relatives of Kaliphora and Melanophylla, the sequence data do not support the relationship proposed by Takhtajan and Thorne for Melanophylla, Kaliphora, and Monlinia. Significantly, Toricellia, Aralidiuin. and Melanophylla all possess leaf sheathes at the base of the petioles, a trait also found in Apiales. These three genera also share empty ovary chambers (Eyde, 1988), a trait not found in Apiales. These shared morphological features therefore agree with rbcL sequence analysis suggesting that these three genera are closely allied with each other, and also the Apiales.
The Phylogenetic analyses of rbcL sequence data identified a strongly supported cornaceous clade that is still present in all trees up to two steps longer than the most parsimonious trees and also has a bootstrap value of 70% (Fig. 2). This cornaceous clade consists of eight of the 17 putative genera of Cornaceae (Alangium, Cornus, Nyssa, Camptotheca, Davidia, Mastixia, Diplopanax, and Curtisia), as well as Hydrangeaceae, and the two sequenced genera of Loasaceae (Petalonyx and Mentzelia). This composition of the rbcL-based cornaceous clade is identical to that of Xiang et al. (1993), except for the addition of Loasaceae, members of which were not analyzed by Xiang et al. (1993). Four well supported major lineages are recognized by the rbcL sequence data, although the relationships among these four groups are not clear: 1) Cornus-Alangium, 2) nyssoids-mastixioids, 3) Curtisia, and 4) Hydrangeaceae-Loasaceae. Genera of the first three lineages, as well as Hydrangeaceae have been allied with Cornus by many authors (Cron quist, 1988; Harms. 1898; Takhtajan, 1987;Thome, 1992) (discussed in Xiang et al., 1993).
The close relationship of Loasaceae to Hydrangeaceae is one of the more significant findings of this study. Loasaceae consist of 15 genera found mainly in tropical and temperate America with one genus in Africa and Arabia. The family has traditionally been considered distantly related to Comus (Cronquist, 1981; Hutchinson, 1967; Takhtajan, 1987; Thorne, 1992). For example, Loasaceae were placed in Dillenii-dae by Cronquist (1981), in Loasales under Loasanae of subclass Dicotyledons by Thorne (1992), in Loasales by Hutchinson (1967), and in Loasales under Loasanae of Lamiidae by Takhtajan (1987). The results of our rbcL sequence analyses clearly indicated that the two genera of Loasaceae (Petalonyx and Mentzelia) included in this analysis are not only members of the cornaceous clade, but most closely related to the hydrangeoids. Some of the shortest trees showed that Loasaceae and Hydrangeaceae are both mono-phyletic and are sister groups, but others showed that Hydrangeaceae are paraphyletic with Hydrangea the sister to Loasaceae. The sister group relationship between Hydrangeaceae and Loasaceae is suggested by more detailed rbcL analysis of all genera of Hydrangeaceae (Soltis et al. in preparation).
Close relationships among Cornus, Hydrangeaceae, and Loasaeeae were recently proposed by Hufford (1990, 1992) and Downie and Palmer (1992). Based on a detailed study of androecial development, Huf ford (1990) proposed that Loasaceae share an ancestry with Cornales and woody saxifrages. The more recent phylogenetic analyses of morphological and chemical data by Hufford (1992) and restriction site mapping of the chloroplast genome inverted repeat by Downie and Palmer (1992) also revealed close relation ships among Comus, Hydrangeaceae, and Loasaceae. Thus our rbcL sequence data agree with the findings of these studies.
As noted above in the introduction, Xiang et al. (1993) revealed that Hydrangeaceae are closely allied with Cornus; hence, in this study we included proposed relatives of Hydrangeaceae to determine whether they too were members of the cornaceous clade. This approach revealed, as noted above, that Loasaceae are part of the cornaceous clade. For this reason we also included Abrophyllum, Quintinia, and Polyosma in
our study, three genera traditionally placed in Escalloniaceae, a family generally considered closely related to Hydrangeaceae (Hutchinson, 1967; Engler, 1890). Abrophyllum and Polyosma have sometimes been treated as separate families (Abrophyllaceae and Polyosmataceae) closely allied with Escalloniaceae (Takhtajan, 1987; Willis, 1973). Our rbcL. sequence analyses revealed, however, that none of these three genera is in the cornaceous clade (Fig. 2). These taxa do not form a monophyletic group with Escallonia. In this analysis, Abrophyllum is allied with Lobelia, Corokia, and Helianthus. More exhaustive phylogenetic analyses of asterids are needed to determine the closest relatives of Abrophyllum. The possible alliance of Abrophyllum with higher asterids is a surprising result, which parallels similar findings for Kaliphora (see the above discussion) and Corokia (Morgan & Soltis, 1993; Olmstead et a., 1993; Xiang et al., 1993).
The phyiogenetic positions of Quintinia, Polyosma, and Escallonia arc uncertain. For example, the strict consensus tree of the 214 trees constituting the first island of minimal length trees shows that Quintinia is a distinct lineage, Polysoma is the sister of Viburnum, and Escallonia is the sister of Berzelia, but these rela tionships were not strongly supported (< 50% bootstrap value). However, the strict consensus tree of the 6 trees constituting the second island of minimal length trees shows that Quintinia is basal to a large clade containing Aucuba, Carrya, and the traditional asterids; Polyosma, Viburnum, and Berzelia form a clade basal to a large clade containing Apiales, Griselinia, Aralidium, Toricellia, and Melanophylla.
Although the relationships among the four genera of Escallomiaceae examined herein remained unre solved, they are clearly not closely related to one another, nor are they close relatives of hydrangeoids based on rbcL sequence data (Fig. 2). Morgan and Soltis (1993) similarly reported based on rbcL sequence analyses that Escallonia and Hydrangeaceae are not closely allied. Ascertaining the affinities of these genera requires additional study of Escalloniaceae, as well as a more exhaustive phyiogenetic analysis of rbcL sequences for Asteridae s. l., such as that conducted by Olmstead et al. (1993).
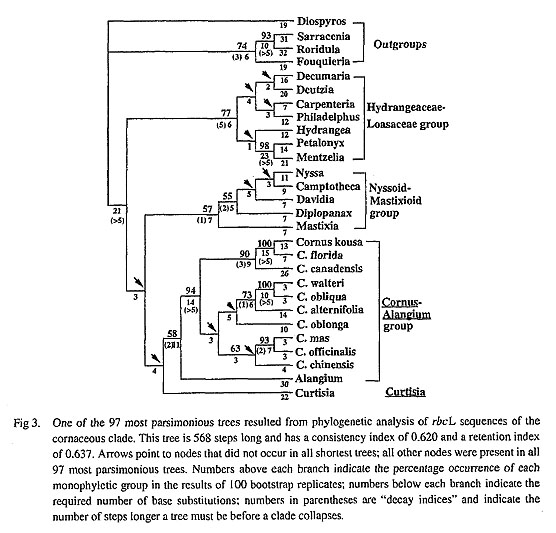
Our analyses of rbcL sequences also have implications for relationships within Cornus, a genus in which relationships have long been debated due to extensive morphological variation (Eyde, 1988; Murrell, 1993; Xiang et at., 1993). The ten sequenced species of Cornus included herein, which represent all of the morphological subgroups of the genus except that the only dioecious species, C. volkensii (African cornelian cherry) and the disciflorous species. C. disciflora (Fig. 3, Fig. 4). Our analyses revealed four major Cornus lineages: 1) Big-bracted dogwoods-Dwarf dogwoods, 2) Cornelian cherries, 3) Blue-fruited (or bractless) dogwoods, and 4) C. oblonga, an unusual, Asian blue-fruited species (see Xiang et al., 1993). These results agree with the earlier findings of Xiang et al. (1993). Furthermore, these groups are very similar to those recognized by Eyde (1988), except that Eyde included C. oblonga in the blue-fruited dogwood lineage and recognized the dwarf dogwoods as a separate major lineage sister to the big-bracted dogwoods within the genus. Relationships among the four lineages detected within Cornus based on rbcL sequences were poorly resolved (< 50% bootstrap values. Fig. 4). Thus, rbcL sequence data provide little support for any of the hypotheses regarding the relative advancement of the red-fruited versus the blue-fruited dogwoods reviewed in Eyde (1988) and Murrell (1993).
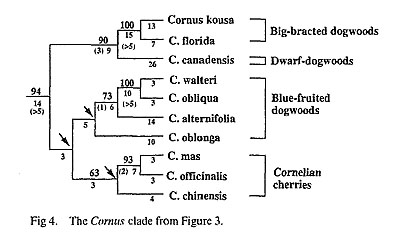
The most recent analysis of morphological characters of Comus (Murrell, 1993) suggested that the big-bracted dogwoods are more closely related to the cornelian cherries than they are to the dwarf dogwoods. Our rbcL sequence data, however, strongly suggest a sister relationship between the big-bracted dogwoods and the dwarf dogwoods (a bootstrap value of 90%, a decay index of 3). Phylogenetic analysis of chloro-plast DNA restriction site data similarly indicates a sister group relation of big-bracted dogwoods and dwarf dogwoods (Xiang et al., 1991). Thus, both of these molecular data sets support the hypothesis of Eyde (1988).
The recent phylogenetic analysis of morphology by Murrell (1993) also suggested a basal position of C oblonga within Comus. Our rbcL sequence data suggest that C. oblonga may be a distinctive lineage within the genus that may be most closely allied with either the blue-fruited dogwoods or cornelian cherries Because there is very little support for any of the basal nodes in this portion of the shortest trees the affini ties of C. oblonga remain uncertain (Fig. 4). An identical duplication near the 3' end of rbcL (see results) was shared by C. oblonga and the sequenced Asian blue-fruited species (C. walteri). This duplication is not present, however, in the North American blue-fruited dogwoods C. obliqua and C. alternifolia. This dupli cation may indicate a sister relationship between the blue-fruited dogwoods and C. oblonga. The duplication may have occurred in the ancestor of these taxa, with subsequent losses in C. obliqua and C. altemifolia. Although, this duplication may have arisen in parallel in C. oblonga and C. walteri.
In summary, this study has again demonstrated that rbcL sequences are valuable in solving systematic problems involving Cornaceae s. l. Our parsimony analyses of rbcL sequence data indicate that Nyssa, Camptotheca, Davidia, Mastixia, Diplopanax, Cornus, Alangium, Curtisia, Hydrangeaceae, and Loasaceae are members of the comaceous clade. This rfccL-based comaceous clade is different in composition from all previously proposed Comus alliances. In contrast, some genera considered members of Cornaceae by some authors, such as Aucuba, Garrya, Corokia, Griselinia, Helwingia, Aralidium, Toricellia, Kaliphora, and Melanophylla are not members of the comaceous clade and only distantly related to Comus. Thus, rbcL sequences indicate that Comaceae s. l. are polyphyletic.
Four major lineages are recognized within this rbcL-based comaceous clade: 1) Nyssoids-Mastixioids, 2) Cornus-Alangium, 3) Curtisia, and 4) Hydrangeaceae-Loasaceae. However, rbcL sequence variation is insufficient to resolve relationships among these four lineages. Sequencing of the chloroplast gene matK, which evolves three times faster than rbcL (Johnson & Soltis, 1994), is being employed to provide greater resolution of phyiogenetic relationships (Xiang et al., ongoing project).
This broad phyiogenetic analysis of rbcL sequences has added a new member, Loasaceae, to the rbcL-based comaceous clade and revealed that four putative genera of Comaceae (Aralidium, Toricellia, Kaliphora, and Menalophylla), as well as three proposed relatives of Hydrangeaceae (Abrophyllum, Quintinia, and Polyosma) are not members of the comaceous clade. Aralidium, Toricellia, and Melanophylla are closely allied with Apiales; Kaliphora, Abrophyllum, Quintinia, and Polyosma are apparently allied with higher asterids. However, the actual affinities of these four genera awaits inclusion in detailed analyses of Asteridae s. l.
Acknowledgments. We thank R. Olmstead for providing an unpublished rbcL sequence of Mentzelia, D. Boufford, E. Wood., I. Hay, S. Yankowski, P. Goldblatt, S. Brunsfeld, J. Wen, H.-N. Qin, Y.-C. Tang, Z.-H. Ji, K.-Y. Pan, K.-Y. Guang, E. Cameron, T. Edwards, E. Oliver, B. Macmillan, A. M. Jayasuriya, P. Gadek, L. Hufford, G. Schatz, Y.-L. Tu, Harvard University Herbaria, Arnold Arboretum, U. S. National
Arboretum, Sirybing Arboretum, Missouri Botanical Garden, and the Royal Botanical Gardens, Kew, England for help in obtaining leaf material. This project was supported by NSF grants BSR 9007614 and DEB 9307000. a WSU travel grant, as well as grants from Sigma Xi. American Society of Plant Taxonomists, and the Northwest Scientific Association.
References
- Adams. J. E. 1949.
- Studies in the comparative anatomy of the Cornaceae. J. Elisha Mitchell Sci. Soc. 65: 218-244.
- Bate-Smith, E. C., I. K. Ferguson, K. Hutson. B. J. Nielsen & T. Swain. 1975.
- Phytochemical interrelationships in the Cornaceae. Biochem. Syst. Ecol. 3: 79-89.
- Chase. M. C., D. E. Soltis. R. G. Olmstead, D. Morgan, D. H. Les, M. R. Duvall. R. Price, H. G. Hills, Y. -L. Qiu, K. A. Kron, J. H. Rettig, E. Conti, J. D. Palmer, M. T. Clegg, J. R. Manhart, K. J. Sytsma, H. J. Michaels, W. J. Kress, M. J. Donoghue, W. D. dark. M. Hedren, B. S. Gaut, R. K. Jansen, K. -J. Kirn, C. F. Wimpee, J. F. Smith, G. R. Furnier, S. H. Straus, Q. -Y. Xiang. G. M. plunkett. P. S. Soltis, L. E. Eguiarte. G. H. Learn, Jr., S. C. H. Barrett. S. Graham & V. A. Albert. 1993.
- Phylogenetics of seed plants: An analysis of nucleotide sequences from the plastid gene rbcL. Ann. Missouri Bot. Gard. 80: 528-580.
- Clegg, M. T. & G. Zurawski. 1992.
- Chloroplast DNA and the study of plant phylogeny: Present status and future prospects. Pp. 1-13. In P. S. Soltis. D. E. Soltis, & J. J. Doyle (editors). Molecular Systematics of Plants. Chapman and Hall, New York.
- Cronquist, A. 1981.
- An Integrated System of Classification of Flowering Plants. Columbia University Press, New York.
- ———. 1988.
- The Evolution and Classification of Flowering Plants, ed.2. New York Botanical Garden Press, New York.
- Dahlgren, R. 1975.
- A system of classification of the angiosperms to be used to demonstrate the distribution of characters. Bot. Not. 128: 119-197.
- ———. 1983.
- General aspects of angiosperm evolution and macrosystematics. Nordic J. Bot. 3: 119-149.
- Donoghue. M. J., R. G. Olmstead, J. F. Smith & J. D. Palmer. 1992.
- Phylogenetic relationships of Dipsacales based on rbcL sequences. Ann. Missouri Bot. Gard. 79: 333-345.
- Downie, S. R. & J. D. Palmer. 1992.
- Restriction site mappings of the chloroplast DNA inverted repeat: a molecular phylogeny of the Asteridae. Ann. Missouri Bot. Gard. 79: 266-283.
- Engler, A. 1890.
- Saxifragaceae. Pp. 42-93. In A. Engler & K. Prantl (editors). Die Naturlichen Pflanzenfamilien, Vol. III. Wilhelm Engelmann, Leipzig.
- Eyde, R- H. 1967.
- The peculiar gynoecial vasculature of Cornaceae and its systematic significance. Phytomorphology 17:172-182.
- ———. 1988.
- Comprehending Cornus: Puzzles and progress in the systematics of the dogwoods, Bot. Rev. 54: 233-351.
- ——— & Q. -Y. Xiang. 1990.
- Fossil mastixioid (Cornaceae) alive in eastern Asia. Amer. J. Bot. 77: 689-692.
- Felsenstein, J. 1985.
- Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791.
- Ferguson, I. K. 1977.
- World Pollen and Spore Flora 6: Cornaceae Dum. Almqvist & Wiksell, Stockholm.
- ——— & M. J. Hideux. 1980.
- Some aspects of the pollen morphology and its taxonomic significance in Cornaceae sensu. lat. Proc. IV. Int. Palynol. Conf., Lucknow 1: 240-249.
- Giannasi. D. E., G. Zurawski & M. T. Clegg. 1992.
- Evolutionary relationships of the Caryophyllidae based on com parative rbcL sequences. Syst. Bot. 17: 1-15.
- Handel-Mazzetti, H. F. V. 1933.
- Plantae novae Chingianae, III. Sinensia 3: 185-198.
- Harms, H. 1898.
- Cornaceae. Pp. 250-270. In A. Engler & K. Prantl (editors). Die Naturlichen Pflamenfamilien, Teil III, Abteilung. W. Engelmann, Leipzig.
- Hufford, L. 1990.
- Androecial development and the problem of monophyly of Loasaceae. Canad. J. Bot. 68: 402-419.
- ———. 1992.
- Rosidae and their relationships to other nonmagnoliid dicotyledons: a phylogenetic analysis using morphological and chemical data. Ann. Missouri Bot. Gard. 79: 218-248.
- Hutchinson, J. 1967.
- The Genera of Flowering Plants. Volume 2. Clarendon Press, Oxford.
- Johnson, L. A. & D. E. Soltis. 1994.
- ORFK DNA sequences and phylogenetic reconstruction in Saxifragaceae sensu stricto. Syst. Bot. 19:143-156.
- Kron, K. A. & M. W. Chase. 1993.
- Systematics of the Ericaceae, Empetraceae, Epacridaceae and related taxa based on rbcL sequence data. Ann. Missouri Bot. Gard. 80: 735-741.
- Li, H. -L. & C. -Y. Chao 1954.
- Comparative anatomy of the woods of the Cornaceae and allies. Quart. J. Taiwan Mus.7: 119-136.
- Linnaeus, C. 1753.
- Species Plantarum. Volume 1, ed. I. London, printed for the Ray Society, sold by Bernard Quaritch Ltd, II Grafton Street London WI. 1957.
- Maddison, D. R. 1991.
- The discovery and importance of multiple islands of most-parsimonious trees. Syst. Zool. 40: 315-328.
- Morgan, D. R. & D. E. Soltis. 1993.
- Phylogenetic relationships among members of Saxifragaceae sensu lato based on rbcL sequence data. Ann. Missouri Bot. Gard. 80; 631-660.
- ———, ——— & K. P. Robertson. 1994.
- Systematic and evolutionary implications of rbcL sequence variation in Rosaceae. Amer. J. Bot. 81: 890-903
- Murrell, Z. E. 1993.
- Phylogenetic relationships in Cornus (Cornaceae). Syst. Bot. 18: 469-495.
- Olmstead, R. G., H. J. Michaels, K. M. Scott & J. D. Palmer. 1992.
- Monophyly of the Asteridae and identification of their major lineages inferred from DNA sequences of rbcL. Ann. Missouri Bot. Gard. 79: 249-265.
- ———, B. Bremer, K. M. Scott & J. D. Palmer. 1993.
- A parsimony analysis of the Asteridae sensu lato based on rbcL sequences. Ann. Missouri Bot. Gard. 80: 700-722.
- Palmer, J. D. 1987.
- Chloroplast DNA evolution and biosystematic uses of chloroplast DNA variation. Amer. Natu ralist 130: S 6-S 29.
- ———, R. K. Jansen, H. J. Michaels, M. W. Chase & J. R. Manhart. 1988.
- Chloroplast DNA variation and plant phylogeny. Ann. Missouri Bot. Gard. 75: 1180-1206.
- Qiu, Y. -L., M. W. Chase, D. H. Les, H. G. Hills & C. R. Parks. 1993.
- Molecular phylogenetics of the Magnoliidae: a cladistic analysis of nucleotide sequences of the plastid gene rbcL. Ann. Missouri Bot. Gard. 80: 587-606.
- Rodman , J., R. Price, K. Karol, E. Conti, K. Sytsma & J. D. Palmer. 1993.
- Nucleotide sequences of the rbcL gene indicate monophyly of mustard oil plants. Ann. Missouri Bot. Gard. 80: 686-699.
- Soltis, D. E., P. S. Soltis, M. T. Clegg & M. Durbin. 1990.
- rbcL sequence divergence and phylogenetic relationships in Saxifragaceae sensu lato. Proc. Natl. Acad. U.S.A. 87: 4640-4644.
- ———, D. R. Morgan, A. Grable, P. S. Soltis & P. Kuzoff. 1993.
- Molecular systematics of Saxifragaceae sensu stricto. Amer. J. Bot. 80: 1056-1081.
- Swofford, D. L. 1993.
- PAUP: phylogenetic analysis using parsimony, version 3.1,1. Laboratory of Molecular Sys-tematics, Smithsonian Institution. Computer program distributed by Center of Biodiversity, Illinois Natural History Survey, Champaign, Illinois.
- Takhtajan, A. L. 1980.
- Outline of the classification of flowering plants (Magnoliophyta). Bot. Rev. 46:225-359.
- ———. 1987.
- System of Magnoliophyta. Academy of Sciences, Leningrad.
- Thorne, R. F. 1983.
- Proposed new realignments in the angiosperms. Nord. J. Bot. 3: 85-117.
- ———. 1992.
- An updated phylogenetic classification of the flowering plants. Aliso 13: 365-389.
- Wangerin, W. 1910.
- Cornaceae. In A. Engler (editor), Das Pflanynreich. Series IV, family 229 (Helf 41). W. Engelmann, Leipzig.
- Willis, J. C. 1973.
- A Dictionary of the Flowering Plants & Ferns. Cambridge University Press, London.
- Xiang, Q. -Y., S. J. Brunsfeld, D. E. Soltis & P. S. Soltis. 1991.
- Molecular systematics of Cornus L. s. l. Amer. J. Bot, 78: 230 (abstract of paper presented at annual meeting).
- ———, D. E. Soltis, D. R. Morgan & P. S. Soltis. 1993.
- Phylogenetic relationships of Comus L. sensu lato and puta tive relatives inferred from rbcL sequence data. Ann. Missouri Bot. Gard. 80:723-734.





